Chapter 5 Taxonomy Profiling
Activity 1 – Trim and QC Reads
Estimated time: 40 min (~25 min computing)
Instructions
1. Import dataset
- Open the GTN Pathogen Detection from Nanopore public history
- Click on
Import this historyand thenCopy History
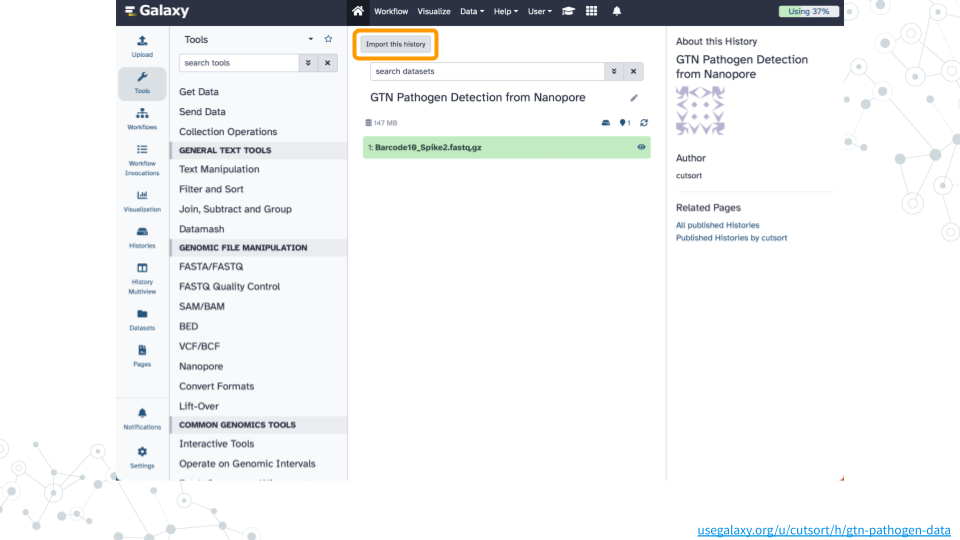
Confirm Barcode10_Spike2.fastq.gz exists in your history by clicking on the Home button in the top menu
2. Run workflow
- Open the Trim and QC Reads public workflow
- Click on
Runand thenRun Workflowon theBarcode10_Spike2.fastq.gzdataset
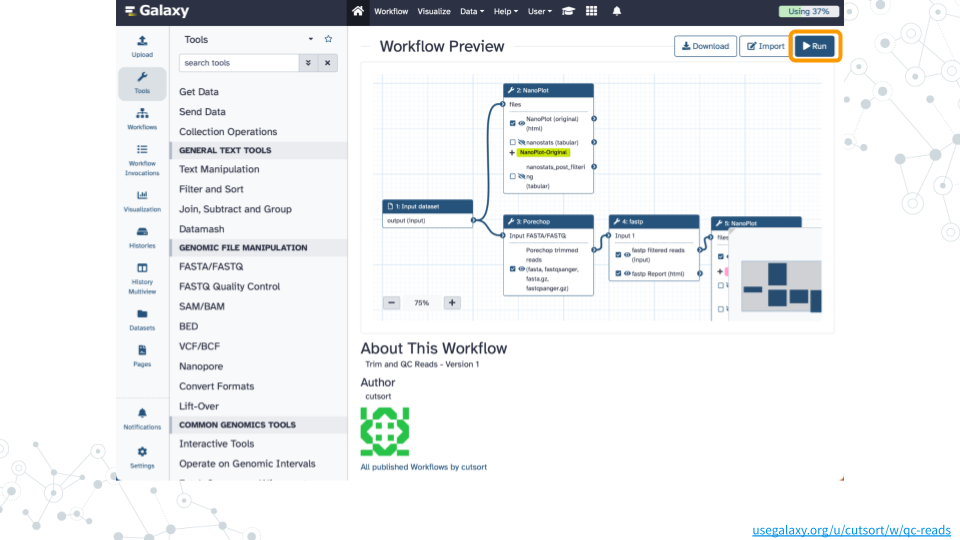
Wait ~25 minutes as the NanoPlot, Porechop, and fastp jobs are scheduled, run, and complete
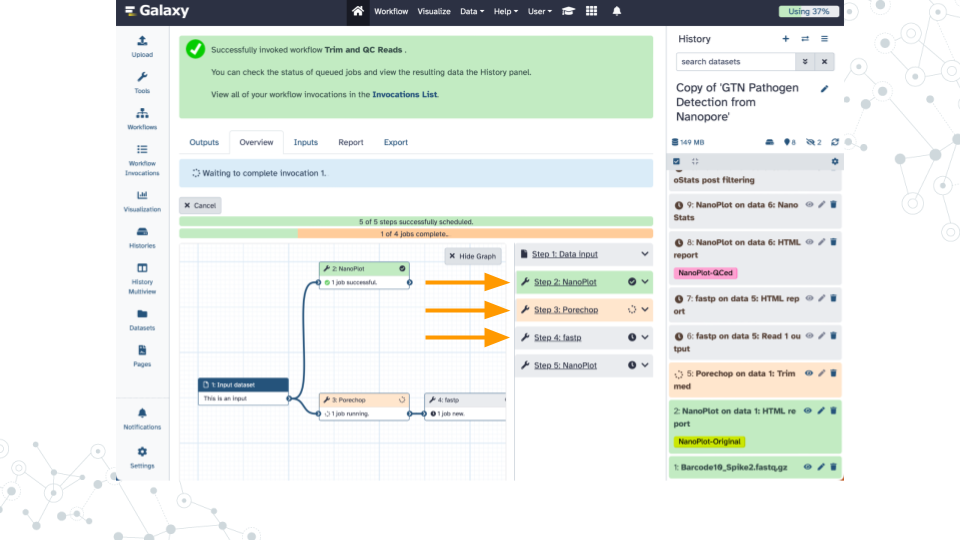
Questions
You can refer to this completed history to answer these questions while you wait for your jobs to complete.
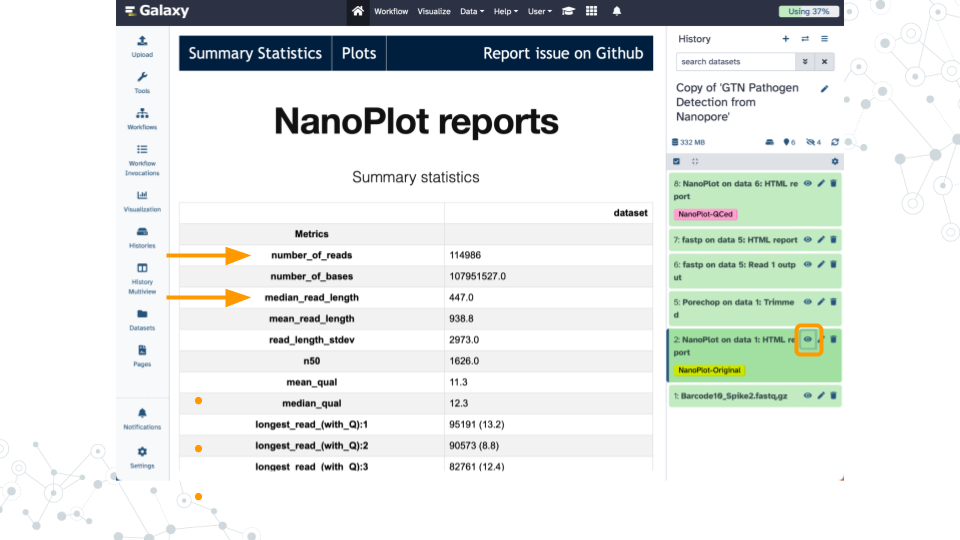
| 1A. How many megabases were sequenced? What percentage was removed by the trimming step? |
|---|
| 1B. What are the median and mean read lengths? Why is the mean is longer than the median? |
|---|
Activity 2 – Taxonomy Profiling
Estimated time: 50 min (~25-35 min computing)
Instructions
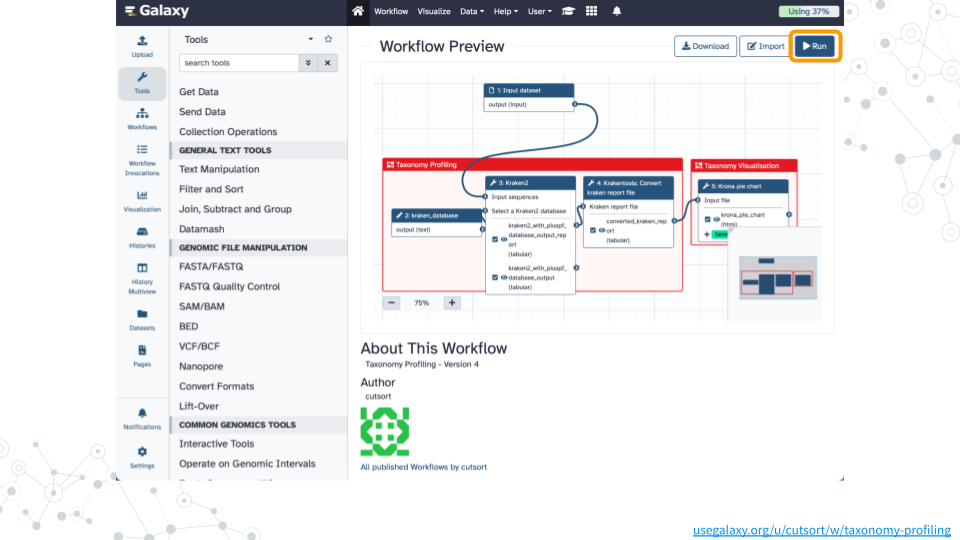
1. Run workflow
- Open the Taxonomy Profiling public workflow
- Click on
Runand thenRun Workflowwith the following parameters- Dataset:
fastp on data 5: Read 1 output - kraken_database:
Prebuilt Refseq indexes: PlusPF
- Dataset:
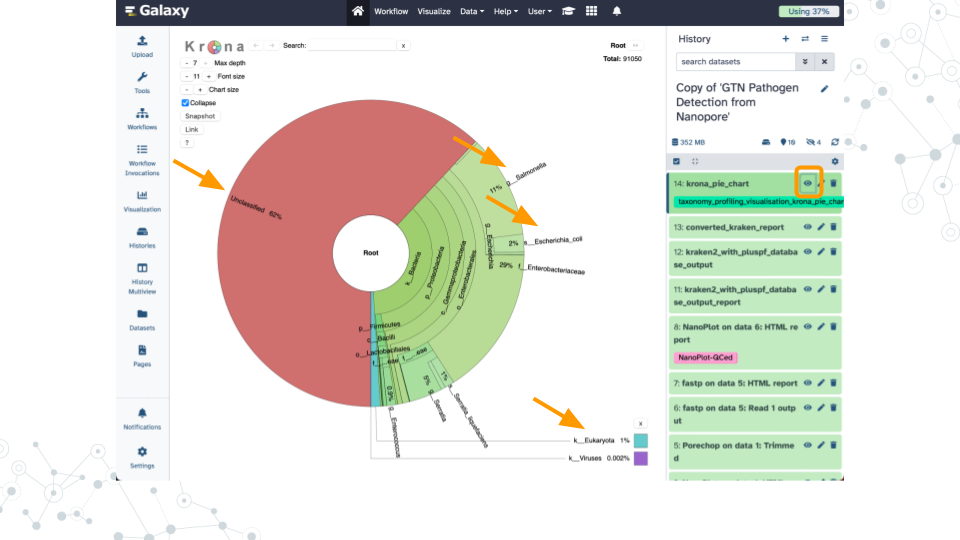
- Wait ~35 minutes as the Kraken2, KrakenTools, and Krona jobs are scheduled, run, and complete
Footnotes
Contributions and Affiliations
- Jennifer Kerr, Notre Dame of Maryland University
- Rosa Alcazar, Clovis Community College
- Frederick Tan, Johns Hopkins University
- Based on “Pathogen detection from (direct Nanopore) sequencing data using Galaxy - Foodborne Edition” (GTN)
Last Revised: September 2024